Unconventional Programming of Embryo Development
Studies like this one are possible because of the public domain datasets available . What we do is similar to assembling a puzzle. We are now given lots of pieces all at once instead of one at a time.”
— Alan Herbert
CHARLESTOWN, MA, UNITED STATES, June 9, 2022 /EINPresswire.com/ -- How can you program a single cell to develop into an organism with many complex feature? Today in a preprint just released on BioRxiv, an international group of scientists provide a
new answer to this long unanswered question. They examine how an embryo bootstraps development through non-classical genetic elements called flipons that target non-coding RNAs to the genes that specify how to make tissues.The classical view of gene regulation is based on studies of bacteria from many decades ago where proteins bind to specific regions of DNA to turn on genes. These approaches have helped ius n many ways to better understand how proteins specify the specialized functions of tissues found in humans and other multi-cellular organism. But is this the full story? How does an embryo start as a single cell and develop into something so complex? How does the embryo know which genes it needs to turn on or off and where they are located in the genome? Is that process fixed at the time of fertilization or determined by some other means?
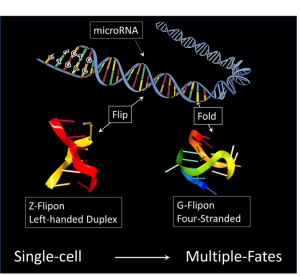
In the manuscript entitled “A role for Flipons and miRNAs in Promoter Specification during Development”, the authors provide evidence that the locations in the genome of the genes involved are marked by flipons, sequences that from alternative DNA structures under physiological conditions. These shapes are different form the right-handed B-DNA helix described by Watson and Crick. They include left-handed helical conformations like Z-DNA, helices with three strands called triplexes and four-stranded folds of DNA into quadruplexes. These genetically encoded elements act as addresses for the genes in which they are located. Their distinctive flipon shapes make these sites easier to find as they stand-out from the billions of bases that make up a human genome. They also help target the binding of regulatory factors to these genes instead of to one of the many thousands of similar looking sequences elsewhere in the genome that are non-functional.
Flipons are enriched in the control region of genes, keeping these regions accessible to factors that guide development. But what are these factors that guide development? The authors find that flipons are close to sequences with matches to microRNAs (miRNAs), a class of small RNAs known previously shown to affect the timing of developmental steps. Once bound, these miRNAs localize enzymes to mark the location of the control elements so that they can be used to control gene expression during development.
The interaction between flipons and miRNAs determines which control elements are marked. It requires a flipon to make a miRNA binding site accessible, while a miRNA that binds an open site is required for a flipon to have any effect. In this way, only a small number of possible miRNA binding sites and flipons are actively engaged in development. Since miRNAs are transmitted by both mother and father, the embryo starts with enough information to mark the genomic locations of genes important for bootstrapping early events. Of course, the nature and quantities of miRNAs transmitted from each parent can vary from embryo to embryo, creating differences in how each one turns out.
The paper brings together previous work on miRNAs with recent studies on the dynamic transformations that occur in DNA structure within a cell. The findings have important implications in how new features can arise over generations. For example, novel miRNA may activate sequences at new genomic locations leading to a different programming of developmental events. The model proposed also provides insights on how to optimize synthetic RNA molecules that regulate gene expression therapeutically.
The scientists draw parallels in their new understanding of embryonic development to the way a computer boots up. On starting, the computer has to initialize its hardware to allow loading of the operating system that controls how inputs are handled. It then runs programs to produce an output. At each step, different types of information are used to specify the system state. Similarly, an embryo needs to initiate the genes that specify how it develops. It needs to flag the genomic locations of genes in a way that they can correctly be turned “on-“ or “off“ as developmental programs vary with the stage of development and with the tissue involved.
Dr. Herbert states that “Studies like the one we report are possible because of the enormous datasets in the public domains that we have access to. What we do is similar to assembling a puzzle. With these datasets, we are given most of the pieces all at once. Previously we would be given them one at a time. With that earlier approach, a lot of time was spent trying to fit pieces together, even when there was not a good match. Now we can do the assembly of the puzzle computationally using snapshots taken from many different vantage points. Then we can check the predictions we can make experimentally to confirm the fit. Of course, we spend most of the time assuming we have made a mistake somewhere in our analysis rather than assuming that everything we did was done with absolute perfection”.
InsideOutBio is an early-stage biotech company developing therapeutics for the treatment of cancer The access to the enormous databases created by collaborative international efforts has helped the InsideOutBio scientists make fundamental discoveries such as those reported by Dr. Herbert, Mr. Pavlov, Mr Konovalov and Dr. Poptsova in the paper. InsideOutBio is privately held. Fedor Pavlov, Dmitrii Konovalov and Maria Poptsova are supported by the Basic Research Program of the National Research University Higher School of Economics, for which Dr. Herbert is an International Supervisor. This research received no external funding

No comments:
Post a Comment